REPORTS
Bench to Bedside: Precision Medicine for Diabetes – A Clinicians Perspective
Presented by:Ananda Basu, MD, FRCP
University of Virginia, Charlottesville, VA; USA
Precision medicine is an emerging approach for disease prevention and treatment that considers individual variability in genes, environment, and lifestyle for each person.1 Precision medicine allows clinicians and researchers to more accurately predict which treatment and prevention strategies will work for a particular disease and in patients with particular characteristics. 1 Although the term “personalized medicine” is sometimes used interchangeably with “precision medicine”, the latter term is preferred because it focuses on the fact that what is being targeted are specific patient characteristics and not the individual per se.
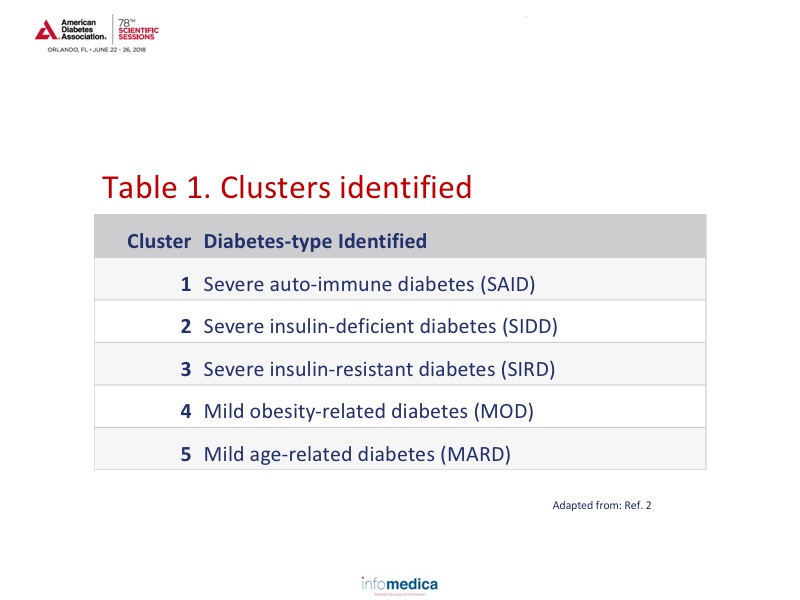
An example of precision medicine study gathering can be seen in the study by Ahlqvist E et al., that focusing on type 2 diabetes clusters.2 A cluster analyses of ~9,000 patients with newly diagnosed type 2 diabetes in Sweden was based on GAD antibodies, BMI, age at diagnosis, HbA1c, HOMA-ß cell function, and HOMA-IR.2 The clusters were related to prospective data from electronic medical records on the development of diabetic complications and medication prescriptions.2 A regression analysis was applied to compare2:
- Time to medication needs
- Time to reach treatment goals
- Diabetic complications
- Genetic associations.
Analyses of the ~9,000 patients identified 5 clusters which can be viewed in Table 1. Based on the cluster analyses over an 8-year period, there appears to be a specific pattern occurring for each of the clusters for HbA1c, age of diagnosis, and HOMA indices.2 Cluster 3 (SIRD) also appears to have an accelerated timeline to chronic kidney disease, macroalbuminuria, end stage renal disease development, and time to cardiovascular disease, a finding that would only be evident through the use of cluster analysis.
Another cluster analysis study focused on insulin resistance and the use of DPP-4 inhibitors and GLP-1 RAs. By focusing on evaluation markers of insulin secretion (C-peptide, urine C-peptide/creatinine ratio, HOMA indices) and action (HOMA-IR, triglycerides, and high density lipoprotein), analyses could be focused on determining the response to DPP-4 inhibitors and GLP-1RAs.3 Findings from the analyses demonstrated that therapy with DPP-4 inhibitors was linked to a lower HbA1c response in patients who had markers of higher insulin resistance.3 There did not appear to be an association between markers of insulin resistance and GLP-1 RA response.3
By combining data from several variables, not simply glucose or HbA1c, more meaningful information should be useful in guiding rational therapeutic strategies for the management of diabetes. In addition, more meaningful diabetes classifications may provide enhanced information.
The genetic markers of metformin pharmacokinetics have shown limited clinical utility in predicting the therapeutic response to metformin. Use of OCT1 inhibitors in those with genetic variation of SLC22A1 led to a 4-fold increase in gastrointestinal adverse effects with metformin use. Unfortunately, the predictive insights and effect size of metformin use have been modest at best, despite considerable progress made in understanding the genetic basis of metformin responsiveness.
Modest progress has also been made in sulfonylureas, in that carriers of TCF7L2 alleles are less likely to reach target HbA1c goals than noncarriers. However, coding variants of sulfonylurea receptors have not been validated sufficiently in prospective studies.
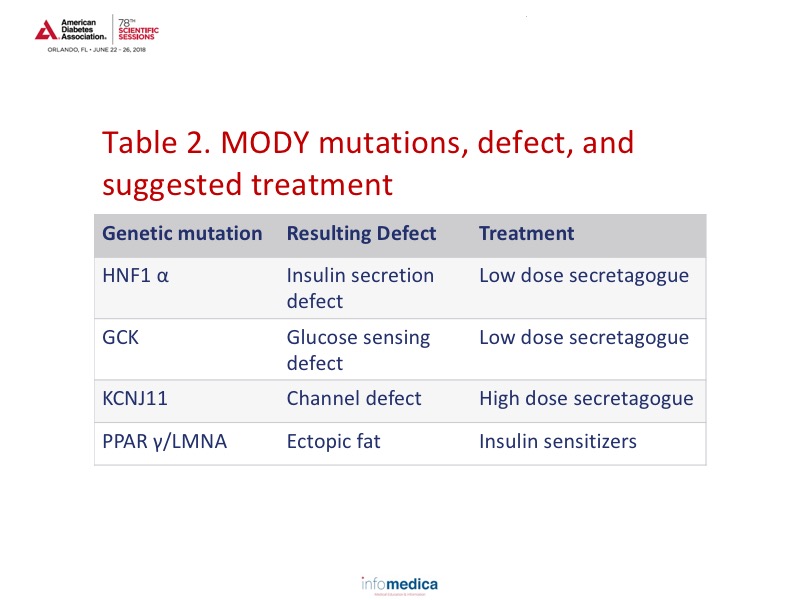
Through precision medicine, information has also been gained on monogenic diabetes (MODY). Approximately 3-4% of children with diabetes are estimated to have MODY. Data has shown which treatments would be more effective in this population based on the specific genetic mutation involved. Review Table 2 for the mutation type, defect type, and suggested treatment.
Precision imaging for diabetes is also available and includes ß cell imaging (pancreas volume and fat, β cell mass), proton magnetic resonance spectroscopy (with 1H-MRS; 18F-SPECT/PET), peripheral tissue imaging, liver imaging (for NAFLD/NASH: MRS, MRI, MRE; US; CT), and cerebral imaging.
Physiologic models of carbohydrate metabolism in diabetes and prediabetes have also been developed for ß cell function and responsivity, insulin sensitivity (peripheral and hepatic), and type 1 diabetes simulator (Food and Drug Administration approved).
Precision simulation takes the approach of creating a simulated virtual image of the patient that includes genomic data, electronic medical records data, physiologic data, and real-time sensing data. By creating a simulated digital twin, recommended medication adjustments and closed-loop control of diabetes can be developed.
Key Messages
- Precision medicine is an emerging approach for disease prevention and treatment that considers individual variability in genes, environment, and lifestyle.
- Cluster analyses can provide valuable information about different specific groups by using multiple parameters as opposed to 1 or 2.
- The pharmacogenetics of diabetes, and specific drug classes in particular, may help lead to more effective treatment and outcomes based on genetic variations.
- Precision imaging, metabolic modeling, and simulation is another approach to provide more focused treatment options and with better outcomes.
REFERENCES
Present disclosure: The presenter reported that he was on an advisory panel for Voluntis and provided research support for Astra Zeneca.
Written by: Debbie Anderson, PhD
Reviewed by: Marco Gallo, MD